sLoLa Workshop 2026
Julie, Deanna, and Nan from the RiboCode team recently attended the BBSRC sLoLa Workshop 2026 in Birmingham. sLoLa (strategic longer and larger) grants from BBSRC are for large team-based fundamental bioscience research projects which push the frontiers of human knowledge. This was a fantastic meeting that brought together teams from across all sLoLa projects to network, exchange knowledge, and further develop best practice in team-based bioscience within the context of longer, larger research projects.
Julie gave a presentation on engaging with new audiences, while Nan presented a poster.
I found this to be a valuable and eye-opening opportunity to learn about the projects of other sLoLa teams and their approaches to leadership, management, and supporting team development. I had engaging discussions with other early career researchers and received constructive feedback and helpful suggestions on my poster. Hearing directly from the funders for the first time was particularly valuable and helped me better understand funding priorities and expectations.
Nan Zhao, postdoctoral researcher
I really enjoyed attending the sLoLa conference, as it was a great opportunity to see the breadth of work across the different sLoLa teams and to learn more about their individual projects. My favourite part of the conference was seeing the many creative and engaging ways in which teams have approached outreach to the public. The range of ideas and formats used to communicate science to undergraduates or to school children was particularly impressive and gave me lots of inspiration for future outreach activities.
Deanna Saunders, PhD student
Thank you to BBSRC for organising a great event.
Research Culture day
We are committed to building a supportive research culture across our multi-institutional, collaborative team. As part of an exciting programme of events to support this, we recently applied for research culture event funding from the University of Leeds – and we were successful!
On 11th July, we came together for an engaging and energising day designed to strengthen the culture across the team outside of our usual work environment. We kicked off with an e-workshop led by Dr Hannah Roberts. Hannah is a scientist, bestselling author and award-winning careers coach focused on helping people – particularly women – working in STEM to fulfil their potential. We opted for Hannah’s signature workshop: ‘The Psychology of Selves: Beyond Imposter Syndrome’, since imposter syndrome is something that we identified as afflicting almost everyone, from PhD students to PIs. To maximise the value of the workshop, we opened this out to the lab groups of the RiboCode PIs, totalling an audience of almost 50. ‘The Psychology of Selves’ was a thought-provoking workshop which coupled light-hearted group discussion with some thoughtful inward reflection. We all came away with a toolkit of practical strategies to start tackling our inner critics.
After lunch, we moved into an afternoon of teambuilding games:
1) Sort Yourself: in small teams, we were tasked with putting ourselves in order according to how we secretly ranked ourselves according to a given metric (e.g. ‘how successful would you have been as a Roman emperor?’, ‘how long can you hold your breath?’, ‘how good are you at drawing?’).

2) Reverse Pictionary: sitting back-to-back, one person gave their partner instructions to draw a picture, without describing the content of the image itself. The aim was for the artist to correctly guess the original image from their masterpiece/disasterpiece!
We managed to include the whole team, including Juan who is pictured here playing Reverse Pictionary with Anton from Spain!
3) Project Lumina: a social deduction game, akin to Werewolves. Working in small teams, we were tasked with uncovering the Alternative Theorists (individuals who believe in a radical hypothesis and subtly steer experiments to generate divergent data that ultimately supports their breakthrough) from the Collaborative Scientists.

To take advantage of the glorious weather, we spent some time relaxing in St George’s Field before heading to the pub for some welcome refreshment. Our teambuilding day reminded us of the value of stepping away from our desks and lab benches to connect with collaborators in a less formal way. We look forward to making this a more regular occurence!

Reflecting on the day’s experiences afterwards, team members reported:
We were encouraged to only share what we were comfortable with – and it really was a safe space. Being open with colleagues really helped us to understand each other’s motivations better.
I got to have deeper conversations with different team members which would not necessarily happen in science-only meetings.
Many thanks to the OD&PL team at the University of Leeds for supporting this event.
Translation UK 2025
Several members of the RiboCode team recently attended Translation UK 2025 in Edinburgh. This was a fantastic conference both for learning about new developments in the field and presenting our own progress on the RiboCode project.
Posters were presented by Veronica Thuburn, Tessa Chan, Jo Cunningham, Nan Zhao, Ella Dimascio, Karl Norris, and Bulat Fatkhullin – Ella even won the award for best student poster! Jo also had the opportunity to chair a session, and Veronica and Ella gave excellent flash talks.

This was my first ever academic conference and, as someone fairly new to the world of translation, it was fantastic to be able to discover the sheer depth of the field and the different applications and angles that come with it. The poster presentations were a particular highlight: the opportunity to discuss cutting-edge research one-on-one was incredibly thought-provoking. I also discovered that I am ‘Team Translon’!
Alex Goncalves, senior research technician
Thanks are due to the organisers and the Biochemical Society for providing a great start to our 2025 conference season.
New paper: Translational activity of 80S monosomes varies dramatically across different tissues
The RiboCode team has recently published a paper that sheds light on the translational activity of ribosomes from various Drosophila melanogaster tissues.
A long-held assumption has been that single ribosomes – known as 80S monosomes – are inactive, while polysomes are actively translating (i.e. producing proteins). However, the team found that the proportion of ribosomes involved in a mRNA translation as 80S monosomes or polysomes varies across cell types and tissues. Specifically, they discovered that 80S monosomes are highly translationally active in the head and embryo, while translationally inactive in the testis and ovary.
Analysis of the mRNAs translated by head 80S monosomes (RNA-Seq) revealed that they preferentially translate mRNAs with specific features and functions. In conclusion, regulation of translation initiation and protein synthesis is mostly performed by monosomes in head and embryo, while polysomes are the main source of protein production in testis and ovary.
“Interestingly, this work developed from an intriguing observation made by a master’s student and a rotation PhD student. We have revealed another contributing factor of protein synthesis regulation and demonstrated that mechanisms of control vary across tissues” – Dr Julie Aspden, senior author
The paper, which is published in Nucleic Acids Research, is available here.
New paper: EMG1 methyltransferase activity affects ribosome occupancy at KSHV uORFs
Members of the RiboCode team have contributed to a recent paper that advances our understanding of how viruses can manipulate host cell translational machinery.
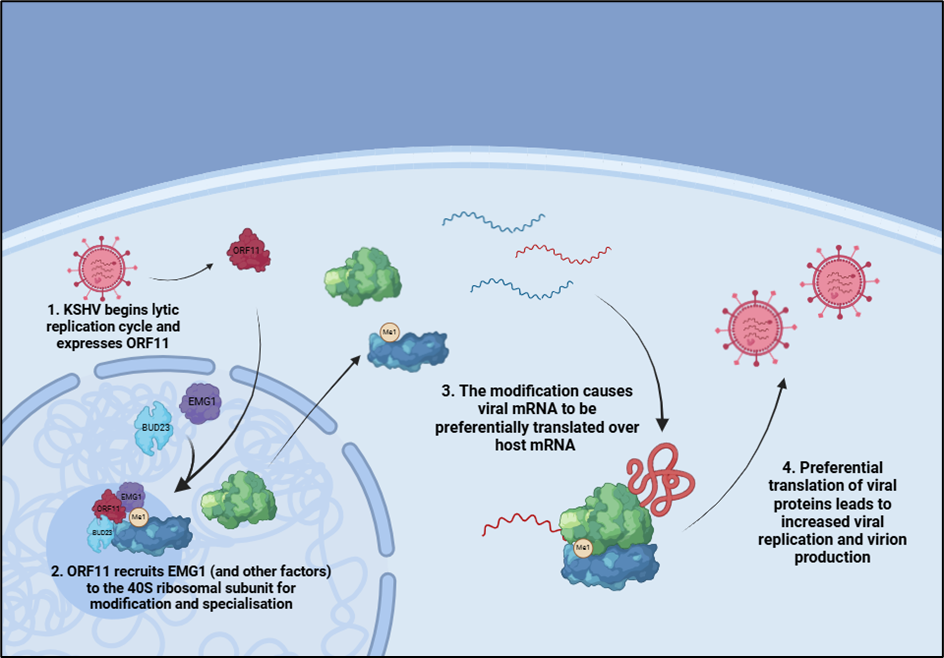
“This is an important first step in our efforts to better understand the molecular mechanisms of how a virus produces specialised ribosomes, it will be exciting to see if similar approaches are conserved in other eukaryote systems”
Professor Ade Whitehouse, senior author
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a human tumour virus associated with several cancers and lymphoproliferative diseases. The authors show that KSHV makes virus-specialised ribosomes by changing the composition of normal host cell ribosomes, thereby allowing the preferential translation of viral transcripts over those of the host.
Building on previous work by the Whitehouse lab, this work also demonstrates the enhanced association of the host protein EMG1 with ribosome subunits during KSHV infection, as well as showing that EMG1 is essential for KSHV lytic replication and infectious virion production. Importantly, the team have shown that the methyltransferase activity of EMG1, which modifies 18S rRNA, is required for enhanced translation of viral mRNAs. This provides evidence that modification of rRNA can affect the activity of ribosomes during infection.
The full paper, published in Cell Reports, is available here.
Special issue and team paper
We have published our first group paper!
Our review on ribosome heterogeneity and specialisation, published in Philosophical Transactions of the Royal Society B, discusses the mechanisms of how specialisation occurs – or might occur – along with a review of the latest research results.
For this paper, we worked together as a cross-disciplinary team to explore the future of ribosome specialisation research, including how technological advancements will drive the next set of research questions, from different perspectives. We propose that examining the evolution of specialisation might help to uncover fundamental principles of translational regulation by the ribosome.
“It was fun to integrate the latest findings, which provide insight into some of the mechanistic details of how translation is regulated, and think about how this could be taken further”
Lead PI of the RiboCode project, Dr Julie Aspden
Our paper is part of a special issue, containing a selection of research articles and reviews covering different aspects of ribosome heterogeneity and specialisation across species, with relevance to human disease. This publication follows up on the Royal Society Hooke meeting in November 2023, and provides an exciting insight into the latest advances and discussions on the future direction of the field.

Our paper, Specialized ribosomes: integrating new insights and current challenges, can be found here. Please contact Dr Julie Aspden for further information.
Credit to RiboCode postdoc Dr Bulat Fatkhullin for the beautiful cover image of an 80S ribosome from the testis of Drosophila melanogaster.
Public engagement activity: Festival of the Mind 2024
Throughout September, some of our RiboCoders had the pleasure of presenting an exhibit at the 7th Festival of the Mind – Sheffield’s biannual collaborative showcase of research and art!
Preparations started at the beginning of the year, when Emma, Veronica and Tess from the Thomson lab in Sheffield reached out to internationally renowned Sheffield-based artist and sculptor Christopher Jarratt. Over a cup of tea, they explained the fundamentals of their research: how ribosomes work, the observation of compositional differences, and the potential functional consequences. In turn, he told them about his previous work, inspirations and motivations. A key theme of Christopher’s work is the use of bold colours and wind-powered kinetic sculptures to visualise complex ideas in a simple and striking way. This really chimed with what the team wanted to do, and so the brainstorming began.
The team wanted to incorporate Christopher’s signature spinning kinetic sculptures to create a 3D animation effect that represented the ratcheting motion of translating ribosomes. Additionally, they wanted to clearly visualise the heterogeneity of ribosomes, which is where Christopher’s experience with the unexpected physics of his art came in. He explained how the placement of the structure with respect to the wind source, as well as the location of any nearby obstacles, can affect the spin speed of each placard in a unique and dynamic way. Our RiboCoders agreed that this was a great way to represent the compositional and functional distinction between different populations of ribosomes.


Over the following months, the RiboCoders met with Christopher regularly to iron out details such as the colour scheme and to ensure that everyone was happy with the design. During that time, they also found out that their exhibit – named “The Ribosome Wind Garden” – would aptly be displayed in the prime location of the Sheffield Winter Garden. It was estimated that an additional 7,000 people visited the garden during the 11-day festival period!
In addition to the exhibit being in place throughout the festival, the collaborators also took part in two interactive events. The first was a public Q&A session, where Christopher and the team discussed a broad range of topics from the origin of their collaboration to the evolutionary history of ribosomes and the RNA world. Some fascinating questions were posed by members of the public, who were keen to learn more about ribosomes! Furthermore, the panel also spoke about the importance of making research accessible and showcasing local culture. The second event was a family-friendly workshop where children had the opportunity to make their own miniature spinning ribosome sculpture.

Overall, the Festival of the Mind experience was an incredibly rewarding one. It certainly wasn’t your typical academic collaboration, but everyone agreed that it gave them fresh perspectives on their own work, and a renewed appreciation for the role played by art and culture in sharing ideas in society.
Public engagement activity: Be Curious 2024
We had a fantastic time at Be Curious – the University of Leeds’ flagship public engagement event – on Saturday 18th May.
Around 180 children visited the RiboCode stall to learn how proteins are made. Visitors matched up custom-made laser-cut DNA and RNA base pairs and used our ribosome wheels to turn their code into a chain of amino acids. Each visitor made a protein keychain to take home, using wooden beads of specific colours to represent their chain of amino acids. The materials for our activity will be made publicly available on our website soon.



We set out to evaluate the effectiveness of our public engagement in terms of perceived knowledge. Using anonymous feedback slips, visitors were asked before and after the activity whether they knew how proteins were made. Happily, after visiting the RiboCode stall 76% visitors asserted that they knew how proteins were made, compared to just 9% beforehand.
The RiboCode Outreach team (Bulat Fatkhullin, Juan Fontana, Tiffany Hicks and Veronica Thuburn) designed and produced the materials for the activity, with Anton Calabrese and Karl Norris assisting with the delivery. Thanks to those who volunteered their time to work on the stall, to the HELIX team at Leeds for their support in producing the materials, and to the Be Curious team for putting on such a brilliant festival!
One year of RiboCode!
This week we celebrated one year of RiboCode! It has been a busy year across the team. As well as an exciting 12 months in the labs, we held our launch event, ran an impact workshop with Dr Jamie Gallagher, kicked off our ECR journal club, delivered several outreach activities and lots more.
Thank you to everyone involved and to BBSRC for their support.
You can now connect with us on LinkedIn.
ECR Journal Club

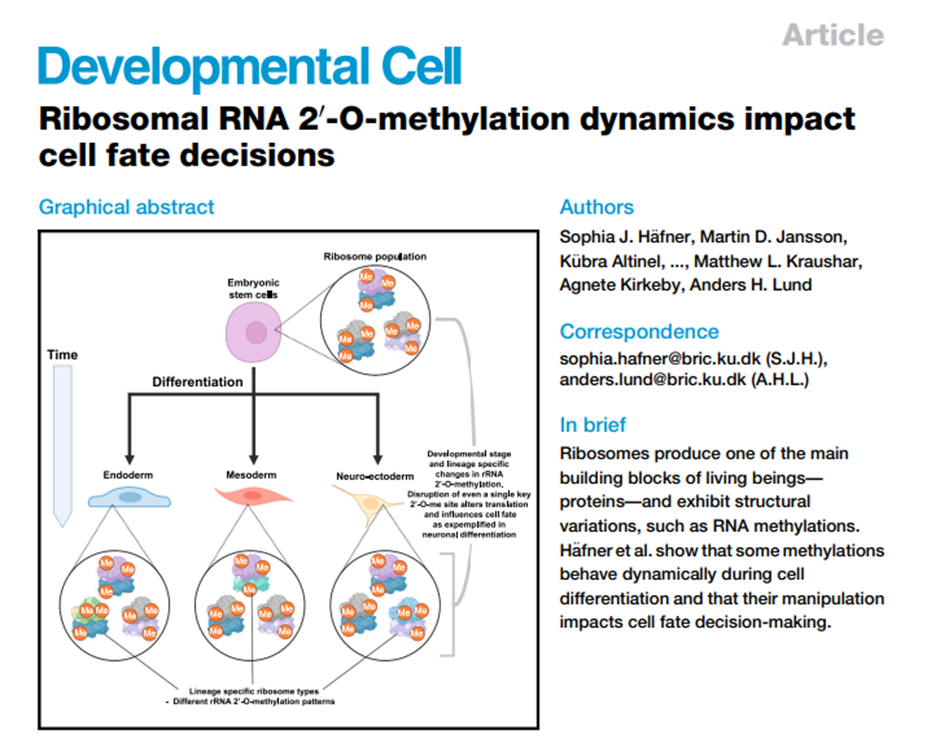
Our fantastic ECR team have established a monthly journal club, taking turns to discuss a paper of interest. This month, Karl Norris led the discussion on this paper by Sophia Häfner and colleagues:
The team then discussed the blurred lines between specialised ribosomes empowered by ribosome-associated factors and RNA-binding proteins recruiting specific mRNAs to the ribosome.
If you have any tips for our budding journal club or any paper suggestions, get in touch!
RiboCode kick-off meeting: Leeds, December 2023

The RiboCode sLoLa has officially launched! On 5th December 2023, the RiboCode team hosted a day of scientific discussion, poster presentations and breakout sessions at the University of Leeds. In attendance was our Scientific Advisory Board (Professors Anne Willis OBE and Alan Warren, both University of Cambridge; Dr Olivia Rissland, University of Colorado, and Dr Mark Bruce, Oxford Nanopore Technologies) and BBSRC Senior Portfolio Manager, Dr Tamsin Shepherd-Waring. We are fortunate to be supported by this inspiring board of advisors and thank them for giving their time and energy to this project.
We were also joined by a number of guests from the University of Leeds, including Simon Wilkins (R&I Development Manager in the Faculty of Engineering and Physical Sciences, who supported the development of the project) and a group of scientists from across the Faculty of Biological Sciences with an interest in the field of specialised ribosomes.
The meeting showcased the work started by our RiboCode scientists, with a real emphasis on the exceptional collaborative nature of the project.
Read more about the RiboCode kick-off meeting here.
Public engagement activity: BioFest 2023
On Wednesday 15th November, our Sheffield scientists – primarily involved in Theme 2 (mRNA translation) – took part in the University of Sheffield’s BioFest Discovery Nights 2023. BioFest is the University of Sheffield’s new bioscience festival, showcasing cutting-edge bioscience research taking part across the university.
Dr Emma Thomson, Dr Jo Cunningham, Vincent Chan and Veronica Thuburn led a fun and engaging activity called ‘Enter the RiboZone’. Here, they used Lego models to how ribosomes decode mRNA and produce protein. Participants took on the role of the ribosome and matched tRNA molecules to an mRNA sequence to spell a word, thus discovering a new appreciation for the complex and essential job of the ribosome in all of our cells!
A whopping 200 visitors visited the stall, with ‘Enter the RiboZone’ proving popular with children of all ages.
Read more about BioFest 2023 here.


Public engagement activity: Be Curious 2023
On Saturday 13th May 2023, members of Theme 3 (structure of specialised ribosomes) took part in Be Curious 2023: the University of Leeds’ annual family open day, which showcases research taking place all over the university. Dr Juan Fontana and colleagues, together with other cryo-electron microscopy experts, led an activity called “Up Close and Really Cold”. Here, they introduced over 200 visitors to cryo-EM sample preparation, freezing samples and studying them under microscopes. The researchers explained how these techniques allow us to understand how diseases can occur in cells and what we can do to intervene.
Read more about Be Curious 2023 here.
BBSRC FTMA-funded visit to Oxford Nanopore Technologies

Three researchers (Elton Vasconcelos, Tayah Hopes and Karl Norris) from the RiboCode team visited Oxford Nanopore Technologies (ONT) for a fortnight in March 2023 to gain a better understanding of an exciting nanopore-based long-read direct RNA sequencing method (ONT dRNA-Seq). They were also keen to discover how research and development was conducted in an industrial setting.
The first week was dedicated to wet lab skills and the second week to bioinformatic analysis. The team learned how to prepare samples for direct RNA sequencing, load MinION flow cells, basecall with Guppy and align to the genome with Minimap2. They also spent time quantifying transcript expression with NanoCount, differential splicing with IsoformSwitchAnalyzeR, and assessing both poly-A tail length and m6A modification with tailFindR and m6anet respectively.
The team wanted to test this technology and perform analyses that would not be possible using next generation sequencing (NGS) technologies. First, they aimed to find out which spliceoforms were being transcribed in their samples. Due to the requirement for RNA to be fragmented to 100bp reads, spliceoform information is often lost during an NGS run. With ONTs long-read sequencing, it is possible to find out exactly which transcript is being sequenced. NGS technologies also require an RNA template to be converted to DNA, losing RNA modification information. With ONTs direct-RNA sequencing methods, it is possible to identify certain RNA modifications. Since the nanopore recognises and initiates sequencing from the PolyA, they were also able to analyse poly-A tail length in different samples.
Samples were run on a MinION chip and achieved read lengths up to 4kb, with a median read length of 1kb. Both the reads’ and bases’ quality fitted above expected (Q score > 7) and, at the end of the full pipeline execution, they were able to identify differentially regulated spliceoforms, different polyA tail lengths and m64A-modified transcripts.
The team were inspired by their industry visit: the technology developed by ONT is hugely exciting and Elton, Tayah and Karl can’t wait to apply what they’ve learned back in Leeds. Thank you to BBSRC for this opportunity and to the ONT team for their warm welcome and hospitality!